|
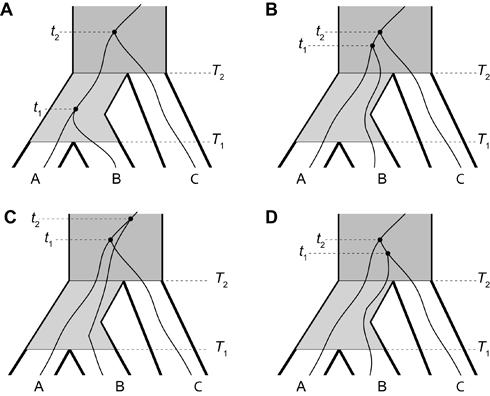
| Three-taxon gene trees within a model three-taxon species tree. The species tree is depicted as a set of heavy black lines and has topology ((AB)C), with species A and B diverging from each other at time T1, and species A and B diverging from species C at time T2. Going backward in time, gene trees are formed by randomly choosing a pair of lineages to coalesce, given that they are in the same population. The first and second coalescent events occur at times t1 and t2, respectively. (A) The gene tree has topology ((AB)C), which matches the species tree. The pair of lineages from species A and B coalesce in the first species that they are capable (i.e., between time T1 and T2). The coalesced lineage of A and B then coalesces with the lineage from C in the species directly ancestral to the root of the species tree (i.e., after time T2). (B, C, D) Due to incomplete lineage sorting, the pair of lineages from A and B fail to coalesce in the ancestral species between time T1 and T2. As a consequence, each pair of lineages from A, B, and C coalesces with equal probability above the root of the species tree. Hence, the gene tree has three possible topologies, which are given by ((AB)C), ((AC)B), and ((BC)A) for parts B, C, and D, respectively. Figure modified from DeGiorgio and Degnan (2010). |